Context and Problem
In scRNA-seq, each cell is sequenced individually, allowing for the analysis of gene expression at the single-cell level. This provides a wealth of information about the cellular identities and states. However, the high dimensionality of the data (thousands of genes) and the technical noise in the data can lead to challenges in accurately clustering the cells. Over-clustering is one such challenge, where cells that are biologically similar are clustered into distinct clusters.
In a previous post, I tested scSHC which tries to solve this problem.
I recently saw this paper A knockoff calibration method to avoid over-clustering in single-cell RNA-sequencing.
The paper introduces a “knockoff calibration method” to address the over-clustering problem. The method involves the use of knockoff features, similar to the approach described earlier, but tailored to the specific challenges of scRNA-seq data.
Knockoff Features Creation: The algorithm generates knockoff features from the original gene expression data. These knockoff features are designed to mimic the original data but are statistically independent of the cell identities or states.
Clustering with Knockoff Features: The algorithm then applies a clustering algorithm to these knockoff features. Since the knockoff features are independent of the cell identities, the clustering should ideally reflect the true structure of the data without being influenced by the biological identities of the cells.
Calibration: The algorithm uses the clustering results from the knockoff features to calibrate the clustering of the original data. This calibration process adjusts the clustering of the original data to avoid over-clustering, ensuring that the final clusters reflect the true biological identities of the cells.
Improved Clustering: By using knockoff features for calibration, the algorithm aims to improve the accuracy of the clustering, reducing the likelihood of over-clustering. This results in more distinct clusters that better reflect the biological diversity of the single-cell population.
The Knockoff Calibration Method implemented in callback R package:
Let’s test it using the same PBMC3k datatset.
library(dplyr)
library(Seurat)
library(scCustomize)
library(patchwork)
library(ggplot2)
library(ComplexHeatmap)
library(SeuratData)
set.seed(1234)data("pbmc3k")
pbmc3k<- UpdateSeuratObject(pbmc3k)
pbmc3k#> An object of class Seurat
#> 13714 features across 2700 samples within 1 assay
#> Active assay: RNA (13714 features, 0 variable features)
#> 2 layers present: counts, dataroutine processing
pbmc3k<- pbmc3k %>%
NormalizeData(normalization.method = "LogNormalize", scale.factor = 10000) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData() %>%
RunPCA(verbose = FALSE) %>%
FindNeighbors(dims = 1:10, verbose = FALSE) %>%
FindClusters(resolution = 0.5, verbose = FALSE) %>%
RunUMAP(dims = 1:10, verbose = FALSE)
p1<- DimPlot_scCustom(pbmc3k, reduction = "umap", label = TRUE, group.by =
"RNA_snn_res.0.5")
p2<- DimPlot_scCustom(pbmc3k, reduction = "umap", label = TRUE, group.by = "seurat_annotations", label.size = 3)
p1 + p2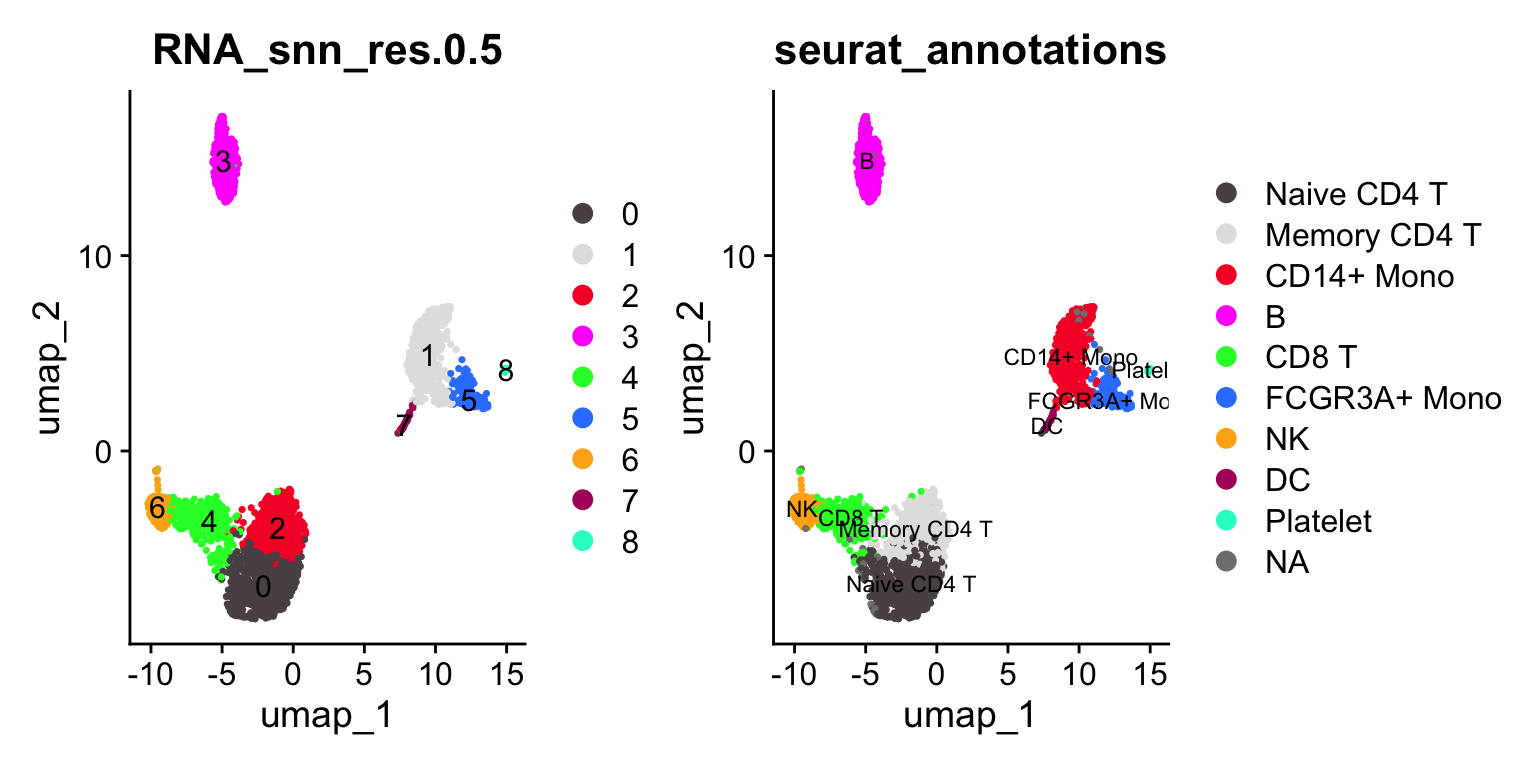
janitor::tabyl(pbmc3k@meta.data, seurat_annotations, RNA_snn_res.0.5)#> seurat_annotations 0 1 2 3 4 5 6 7 8
#> Naive CD4 T 608 0 66 0 23 0 0 0 0
#> Memory CD4 T 69 0 396 0 16 0 2 0 0
#> CD14+ Mono 0 472 0 0 0 3 0 4 1
#> B 0 0 0 343 1 0 0 0 0
#> CD8 T 2 0 3 0 265 0 1 0 0
#> FCGR3A+ Mono 0 7 0 0 0 155 0 0 0
#> NK 0 0 0 0 16 0 139 0 0
#> DC 0 0 0 0 0 0 0 32 0
#> Platelet 0 0 0 0 0 0 0 0 14
#> <NA> 17 12 2 6 18 1 6 0 0Let’s artificially increase the resolution (to 1) to over-cluster it.
## artificially increase the resolution
pbmc3k<- pbmc3k %>%
FindNeighbors(dims = 1:10, verbose = FALSE) %>%
FindClusters(resolution = 1, verbose = FALSE)
p3<- DimPlot_scCustom(pbmc3k, reduction = "umap", label = TRUE, group.by = "RNA_snn_res.1")
(p1 + p3) / p2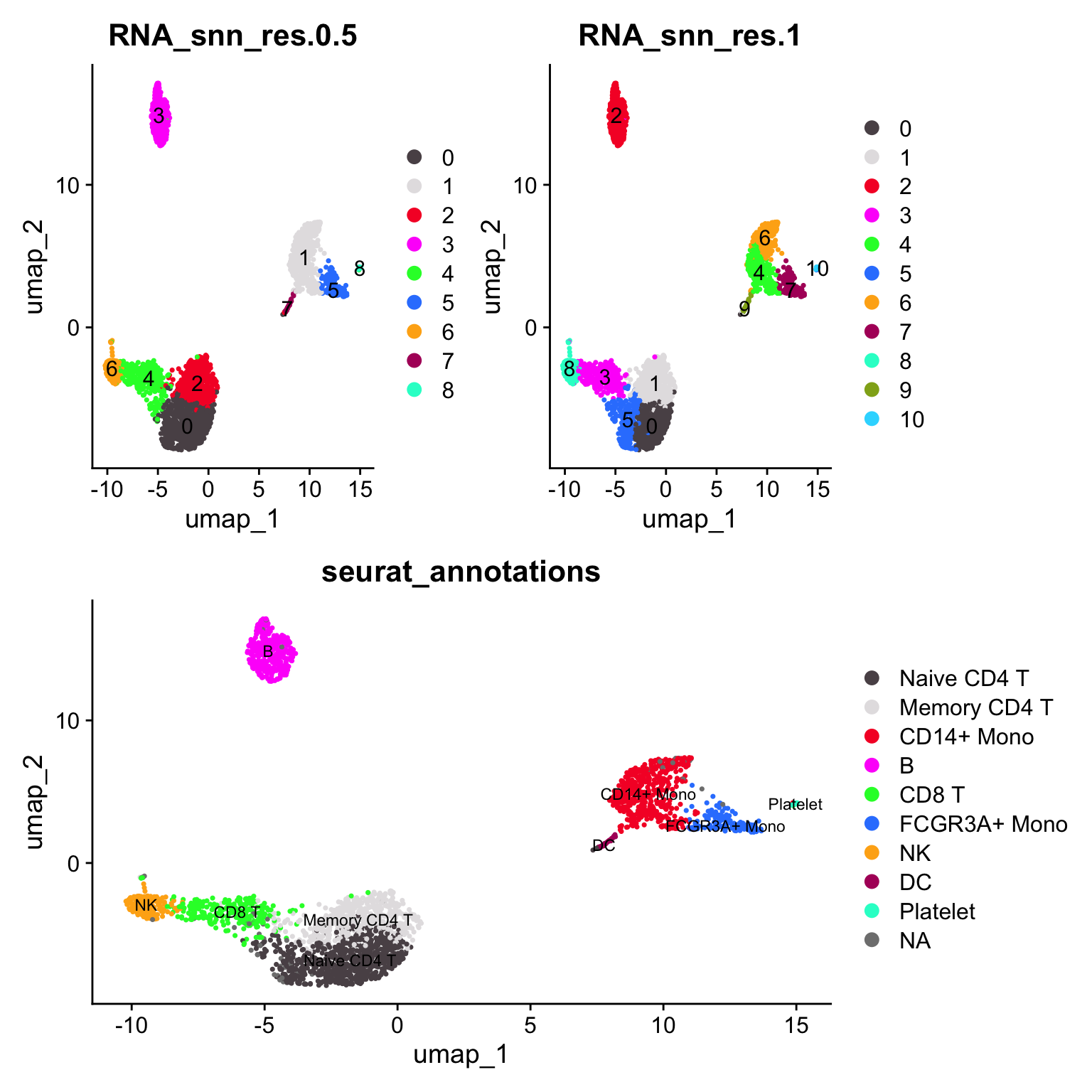
CD4 naive cell cluster is split to 2 clusters (0 -> 0, 5) the CD14+ monocyte cluster is split into 2 clusters (1 -> 4, 6)
janitor::tabyl(pbmc3k@meta.data, RNA_snn_res.1, RNA_snn_res.0.5) #> RNA_snn_res.1 0 1 2 3 4 5 6 7 8
#> 0 471 0 9 0 0 0 0 0 0
#> 1 4 0 455 0 0 0 0 0 0
#> 2 0 0 0 349 0 0 0 0 0
#> 3 0 0 2 0 303 0 0 0 0
#> 4 0 268 0 0 0 1 0 0 0
#> 5 221 0 1 0 36 0 0 0 0
#> 6 0 223 0 0 0 0 0 0 0
#> 7 0 0 0 0 0 158 0 0 0
#> 8 0 0 0 0 0 0 148 0 0
#> 9 0 0 0 0 0 0 0 36 0
#> 10 0 0 0 0 0 0 0 0 15visualize it using https://github.com/crazyhottommy/scclusteval
library(scclusteval)
PairWiseJaccardSetsHeatmap(pbmc3k$RNA_snn_res.0.5,
pbmc3k$RNA_snn_res.1,
show_row_dend = F, show_column_dend = F,
cluster_row = F, cluster_column =F)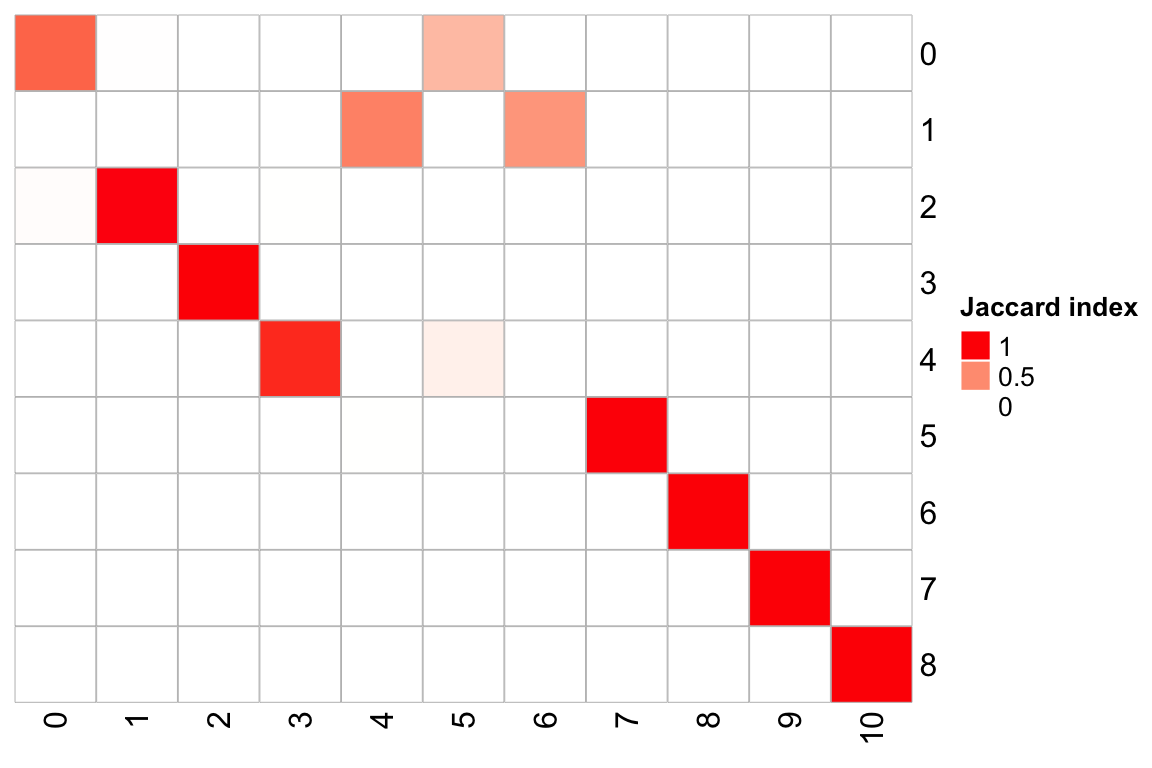
Test callback
Install it here https://github.com/lcrawlab/callback
#devtools::install_github("lcrawlab/callback")
library(callback)
library(tictoc)
tic()
pbmc3k_callback <- FindClustersCallback(pbmc3k)
toc()#> 53.227 sec elapsedp4<- scCustomize::DimPlot_scCustom(pbmc3k_callback)
(p1 + p4) / p2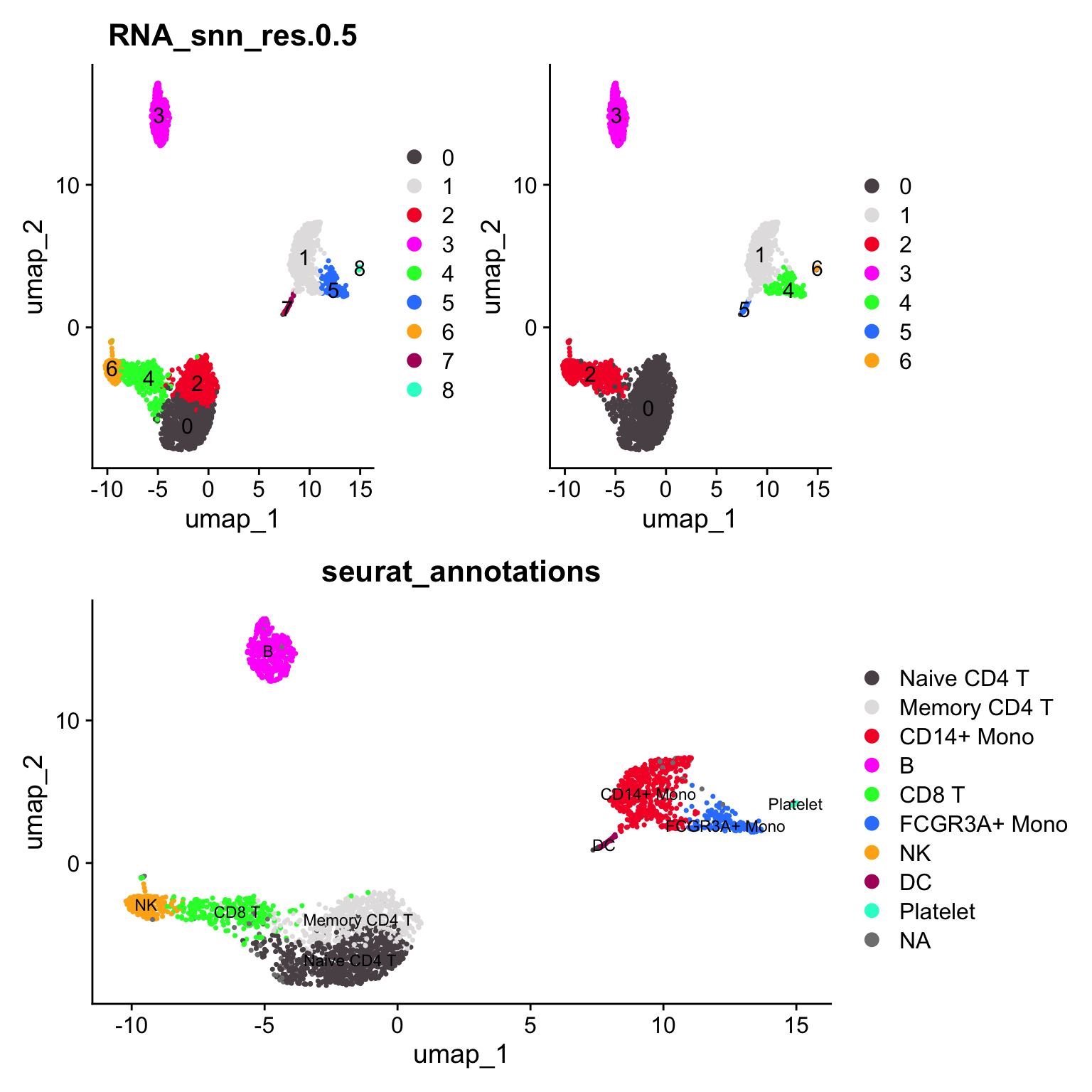
PairWiseJaccardSetsHeatmap(pbmc3k$seurat_annotations,
pbmc3k_callback$callback_clusters,
show_row_dend = F, show_column_dend = F,
cluster_row = F, cluster_column =F)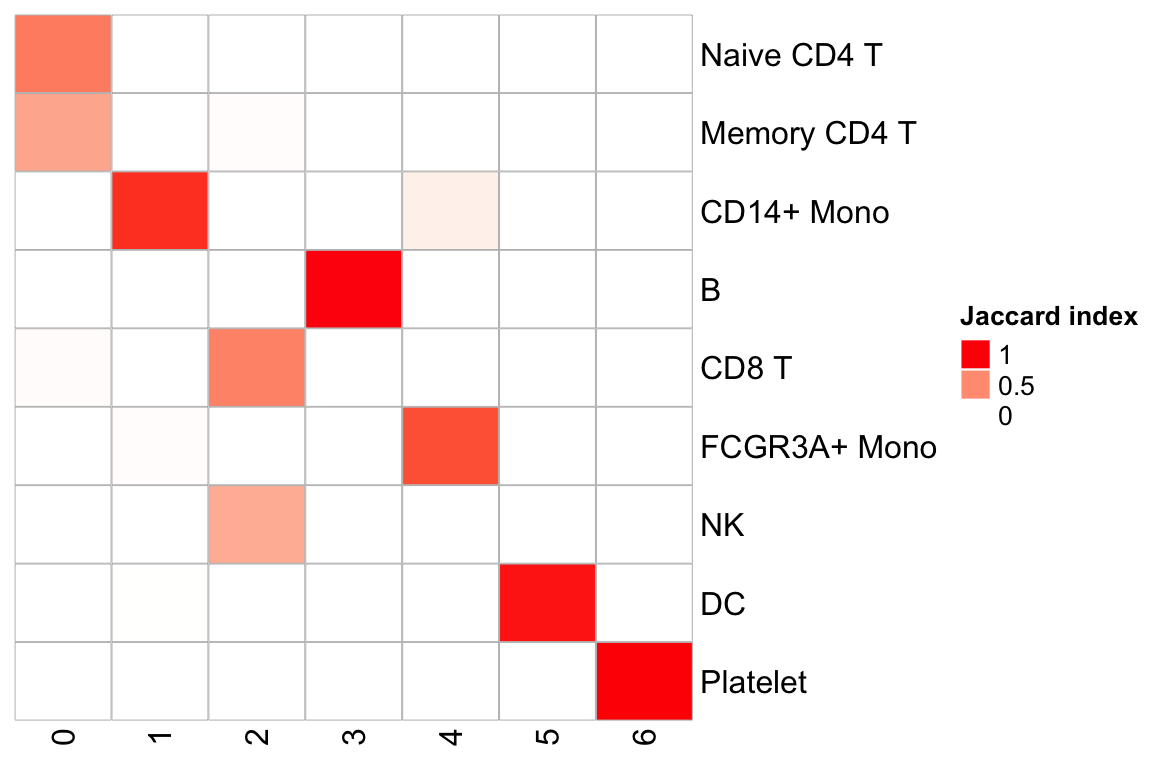
callback merges the CD8 T and NK cells into cluster 2, and merges the
naive CD4 T cells and the memory CD4 T cells. Of course, you do not know the
original seurat cluster annotation is 100% correct or not. Distinguishing naive
and memory CD4 is harder too. However, merging NK cells with CD8T cells do make me worry :)
conclusions
Similar to
scSHC, while statistically attractive, we need to use it with precaution and validate the clusters with biology knowledge.Always use a small dataset that you are familiar with to test a new method.
The cluster number will be depending on the parameter
dims = 1:10in theFindClustersCallbackfunction too.I may still cluster with a bunch of different resolutions and make sense of them using biology knowledge and manually merge or split the clusters.
what’s your take?
devtools::session_info()#> ─ Session info 😕 🤕 📮 ─────────────────────────────────────────────────
#> hash: confused face, face with head-bandage, postbox
#>
#> setting value
#> version R version 4.1.2 (2021-11-01)
#> os macOS Big Sur 10.16
#> system x86_64, darwin17.0
#> ui X11
#> language (EN)
#> collate en_US.UTF-8
#> ctype en_US.UTF-8
#> tz America/New_York
#> date 2024-03-22
#> pandoc 3.1.1 @ /Applications/RStudio.app/Contents/Resources/app/quarto/bin/tools/ (via rmarkdown)
#>
#> ─ Packages ───────────────────────────────────────────────────────────────────
#> package * version date (UTC) lib source
#> abind 1.4-5 2016-07-21 [1] CRAN (R 4.1.0)
#> beeswarm 0.4.0 2021-06-01 [1] CRAN (R 4.1.0)
#> BiocGenerics 0.40.0 2021-10-26 [1] Bioconductor
#> blogdown 1.7 2021-12-19 [1] CRAN (R 4.1.2)
#> bookdown 0.24 2021-09-02 [1] CRAN (R 4.1.0)
#> bslib 0.3.1 2021-10-06 [1] CRAN (R 4.1.0)
#> cachem 1.0.6 2021-08-19 [1] CRAN (R 4.1.0)
#> callback * 0.0.0 2024-03-15 [1] Github (lcrawlab/callback@655705f)
#> callr 3.7.0 2021-04-20 [1] CRAN (R 4.1.0)
#> circlize 0.4.13 2021-06-09 [1] CRAN (R 4.1.0)
#> cli 3.6.1 2023-03-23 [1] CRAN (R 4.1.2)
#> clue 0.3-60 2021-10-11 [1] CRAN (R 4.1.0)
#> cluster 2.1.2 2021-04-17 [1] CRAN (R 4.1.2)
#> codetools 0.2-18 2020-11-04 [1] CRAN (R 4.1.2)
#> colorspace 2.0-2 2021-06-24 [1] CRAN (R 4.1.0)
#> ComplexHeatmap * 2.10.0 2021-10-26 [1] Bioconductor
#> cowplot 1.1.1 2020-12-30 [1] CRAN (R 4.1.0)
#> crayon 1.4.2 2021-10-29 [1] CRAN (R 4.1.0)
#> data.table 1.14.2 2021-09-27 [1] CRAN (R 4.1.0)
#> DBI 1.1.1 2021-01-15 [1] CRAN (R 4.1.0)
#> deldir 1.0-6 2021-10-23 [1] CRAN (R 4.1.0)
#> desc 1.4.0 2021-09-28 [1] CRAN (R 4.1.0)
#> devtools 2.4.2 2021-06-07 [1] CRAN (R 4.1.0)
#> digest 0.6.28 2021-09-23 [1] CRAN (R 4.1.0)
#> doParallel 1.0.17 2022-02-07 [1] CRAN (R 4.1.2)
#> dotCall64 1.1-1 2023-11-28 [1] CRAN (R 4.1.2)
#> dplyr * 1.1.2 2023-04-20 [1] CRAN (R 4.1.2)
#> ellipsis 0.3.2 2021-04-29 [1] CRAN (R 4.1.0)
#> evaluate 0.14 2019-05-28 [1] CRAN (R 4.1.0)
#> fansi 0.5.0 2021-05-25 [1] CRAN (R 4.1.0)
#> farver 2.1.0 2021-02-28 [1] CRAN (R 4.1.0)
#> fastDummies 1.7.3 2023-07-06 [1] CRAN (R 4.1.2)
#> fastmap 1.1.0 2021-01-25 [1] CRAN (R 4.1.0)
#> fitdistrplus 1.1-6 2021-09-28 [1] CRAN (R 4.1.0)
#> forcats 0.5.1 2021-01-27 [1] CRAN (R 4.1.0)
#> foreach 1.5.1 2020-10-15 [1] CRAN (R 4.1.0)
#> fs 1.5.0 2020-07-31 [1] CRAN (R 4.1.0)
#> future 1.25.0 2022-04-24 [1] CRAN (R 4.1.2)
#> future.apply 1.8.1 2021-08-10 [1] CRAN (R 4.1.0)
#> generics 0.1.3 2022-07-05 [1] CRAN (R 4.1.2)
#> GetoptLong 1.0.5 2020-12-15 [1] CRAN (R 4.1.0)
#> ggbeeswarm 0.6.0 2017-08-07 [1] CRAN (R 4.1.0)
#> ggplot2 * 3.4.4 2023-10-12 [1] CRAN (R 4.1.2)
#> ggprism 1.0.3.9000 2021-12-07 [1] Github (csdaw/ggprism@e21c3ee)
#> ggrastr 1.0.1 2021-12-08 [1] CRAN (R 4.1.0)
#> ggrepel 0.9.3 2023-02-03 [1] CRAN (R 4.1.2)
#> ggridges 0.5.4 2022-09-26 [1] CRAN (R 4.1.2)
#> GlobalOptions 0.1.2 2020-06-10 [1] CRAN (R 4.1.0)
#> globals 0.14.0 2020-11-22 [1] CRAN (R 4.1.0)
#> glue 1.6.2 2022-02-24 [1] CRAN (R 4.1.2)
#> goftest 1.2-3 2021-10-07 [1] CRAN (R 4.1.0)
#> gridExtra 2.3 2017-09-09 [1] CRAN (R 4.1.0)
#> gtable 0.3.0 2019-03-25 [1] CRAN (R 4.1.0)
#> highr 0.9 2021-04-16 [1] CRAN (R 4.1.0)
#> htmltools 0.5.2 2021-08-25 [1] CRAN (R 4.1.0)
#> htmlwidgets 1.5.4 2021-09-08 [1] CRAN (R 4.1.0)
#> httpuv 1.6.3 2021-09-09 [1] CRAN (R 4.1.0)
#> httr 1.4.2 2020-07-20 [1] CRAN (R 4.1.0)
#> ica 1.0-2 2018-05-24 [1] CRAN (R 4.1.0)
#> ifnb.SeuratData * 3.1.0 2024-01-17 [1] local
#> igraph 1.2.7 2021-10-15 [1] CRAN (R 4.1.0)
#> IRanges 2.28.0 2021-10-26 [1] Bioconductor
#> irlba 2.3.5.1 2022-10-03 [1] CRAN (R 4.1.2)
#> iterators 1.0.13 2020-10-15 [1] CRAN (R 4.1.0)
#> janitor 2.1.0 2021-01-05 [1] CRAN (R 4.1.0)
#> jquerylib 0.1.4 2021-04-26 [1] CRAN (R 4.1.0)
#> jsonlite 1.7.2 2020-12-09 [1] CRAN (R 4.1.0)
#> KernSmooth 2.23-20 2021-05-03 [1] CRAN (R 4.1.2)
#> knitr 1.36 2021-09-29 [1] CRAN (R 4.1.0)
#> knockoff 0.3.6 2022-08-15 [1] CRAN (R 4.1.2)
#> labeling 0.4.2 2020-10-20 [1] CRAN (R 4.1.0)
#> lamW 2.2.3 2023-12-01 [1] CRAN (R 4.1.2)
#> later 1.3.0 2021-08-18 [1] CRAN (R 4.1.0)
#> lattice 0.20-45 2021-09-22 [1] CRAN (R 4.1.2)
#> lazyeval 0.2.2 2019-03-15 [1] CRAN (R 4.1.0)
#> leiden 0.3.9 2021-07-27 [1] CRAN (R 4.1.0)
#> lifecycle 1.0.3 2022-10-07 [1] CRAN (R 4.1.2)
#> limma 3.50.0 2021-10-26 [1] Bioconductor
#> listenv 0.8.0 2019-12-05 [1] CRAN (R 4.1.0)
#> lmtest 0.9-39 2021-11-07 [1] CRAN (R 4.1.0)
#> lubridate 1.8.0 2021-10-07 [1] CRAN (R 4.1.0)
#> magick 2.7.4 2023-03-09 [1] CRAN (R 4.1.2)
#> magrittr 2.0.1 2020-11-17 [1] CRAN (R 4.1.0)
#> MASS 7.3-54 2021-05-03 [1] CRAN (R 4.1.2)
#> Matrix 1.6-3 2023-11-14 [1] CRAN (R 4.1.2)
#> matrixStats 0.61.0 2021-09-17 [1] CRAN (R 4.1.0)
#> memoise 2.0.0 2021-01-26 [1] CRAN (R 4.1.0)
#> mime 0.12 2021-09-28 [1] CRAN (R 4.1.0)
#> miniUI 0.1.1.1 2018-05-18 [1] CRAN (R 4.1.0)
#> munsell 0.5.0 2018-06-12 [1] CRAN (R 4.1.0)
#> nlme 3.1-153 2021-09-07 [1] CRAN (R 4.1.2)
#> paletteer 1.4.0 2021-07-20 [1] CRAN (R 4.1.0)
#> parallelly 1.31.1 2022-04-22 [1] CRAN (R 4.1.2)
#> patchwork * 1.1.1 2020-12-17 [1] CRAN (R 4.1.0)
#> pbapply 1.5-0 2021-09-16 [1] CRAN (R 4.1.0)
#> pbmc3k.SeuratData * 3.1.4 2022-08-03 [1] local
#> pbmcref.SeuratData * 1.0.0 2023-09-14 [1] local
#> pillar 1.9.0 2023-03-22 [1] CRAN (R 4.1.2)
#> pkgbuild 1.2.0 2020-12-15 [1] CRAN (R 4.1.0)
#> pkgconfig 2.0.3 2019-09-22 [1] CRAN (R 4.1.0)
#> pkgload 1.2.3 2021-10-13 [1] CRAN (R 4.1.0)
#> plotly 4.10.0 2021-10-09 [1] CRAN (R 4.1.0)
#> plyr 1.8.6 2020-03-03 [1] CRAN (R 4.1.0)
#> png 0.1-8 2022-11-29 [1] CRAN (R 4.1.2)
#> polyclip 1.10-0 2019-03-14 [1] CRAN (R 4.1.0)
#> presto 1.0.0 2023-03-30 [1] Github (immunogenomics/presto@045390a)
#> prettyunits 1.1.1 2020-01-24 [1] CRAN (R 4.1.0)
#> prismatic 1.1.0 2021-10-17 [1] CRAN (R 4.1.0)
#> processx 3.5.2 2021-04-30 [1] CRAN (R 4.1.0)
#> progressr 0.9.0 2021-09-24 [1] CRAN (R 4.1.0)
#> promises 1.2.0.1 2021-02-11 [1] CRAN (R 4.1.0)
#> ps 1.6.0 2021-02-28 [1] CRAN (R 4.1.0)
#> purrr 1.0.1 2023-01-10 [1] CRAN (R 4.1.2)
#> R6 2.5.1 2021-08-19 [1] CRAN (R 4.1.0)
#> RANN 2.6.1 2019-01-08 [1] CRAN (R 4.1.0)
#> rappdirs 0.3.3 2021-01-31 [1] CRAN (R 4.1.0)
#> RColorBrewer 1.1-2 2014-12-07 [1] CRAN (R 4.1.0)
#> Rcpp 1.0.11 2023-07-06 [1] CRAN (R 4.1.2)
#> RcppAnnoy 0.0.19 2021-07-30 [1] CRAN (R 4.1.0)
#> RcppHNSW 0.3.0 2020-09-06 [1] CRAN (R 4.1.0)
#> RcppParallel 5.1.4 2021-05-04 [1] CRAN (R 4.1.0)
#> rematch2 2.1.2 2020-05-01 [1] CRAN (R 4.1.0)
#> remotes 2.4.1 2021-09-29 [1] CRAN (R 4.1.0)
#> reshape2 1.4.4 2020-04-09 [1] CRAN (R 4.1.0)
#> reticulate 1.28 2023-01-27 [1] CRAN (R 4.1.2)
#> rjson 0.2.20 2018-06-08 [1] CRAN (R 4.1.0)
#> rlang 1.1.3 2024-01-10 [1] CRAN (R 4.1.2)
#> rmarkdown 2.11 2021-09-14 [1] CRAN (R 4.1.0)
#> ROCR 1.0-11 2020-05-02 [1] CRAN (R 4.1.0)
#> rprojroot 2.0.2 2020-11-15 [1] CRAN (R 4.1.0)
#> RSpectra 0.16-0 2019-12-01 [1] CRAN (R 4.1.0)
#> rstudioapi 0.13 2020-11-12 [1] CRAN (R 4.1.0)
#> Rtsne 0.15 2018-11-10 [1] CRAN (R 4.1.0)
#> S4Vectors 0.32.2 2021-11-07 [1] Bioconductor
#> sass 0.4.0 2021-05-12 [1] CRAN (R 4.1.0)
#> scales 1.3.0 2023-11-28 [1] CRAN (R 4.1.2)
#> scattermore 1.2 2023-06-12 [1] CRAN (R 4.1.2)
#> scclusteval * 0.0.0.9000 2022-08-05 [1] Github (crazyhottommy/scclusteval@b1b22c7)
#> scCustomize * 2.1.2 2024-02-28 [1] CRAN (R 4.1.2)
#> sctransform 0.4.1 2023-10-19 [1] CRAN (R 4.1.2)
#> sessioninfo 1.2.1 2021-11-02 [1] CRAN (R 4.1.0)
#> Seurat * 5.0.1 2023-11-17 [1] CRAN (R 4.1.2)
#> SeuratData * 0.2.2 2022-08-03 [1] Github (satijalab/seurat-data@d6a8ce6)
#> SeuratObject * 5.0.1 2023-11-17 [1] CRAN (R 4.1.2)
#> shape 1.4.6 2021-05-19 [1] CRAN (R 4.1.0)
#> shiny 1.7.1 2021-10-02 [1] CRAN (R 4.1.0)
#> snakecase 0.11.0 2019-05-25 [1] CRAN (R 4.1.0)
#> sp * 1.6-0 2023-01-19 [1] CRAN (R 4.1.2)
#> spam 2.10-0 2023-10-23 [1] CRAN (R 4.1.2)
#> spatstat.data 3.0-0 2022-10-21 [1] CRAN (R 4.1.2)
#> spatstat.explore 3.0-6 2023-01-26 [1] CRAN (R 4.1.2)
#> spatstat.geom 3.0-6 2023-01-30 [1] CRAN (R 4.1.2)
#> spatstat.random 3.1-3 2023-01-25 [1] CRAN (R 4.1.2)
#> spatstat.sparse 3.0-0 2022-10-21 [1] CRAN (R 4.1.2)
#> spatstat.utils 3.0-1 2022-10-19 [1] CRAN (R 4.1.2)
#> stringi 1.7.5 2021-10-04 [1] CRAN (R 4.1.0)
#> stringr 1.5.1 2023-11-14 [1] CRAN (R 4.1.2)
#> survival 3.2-13 2021-08-24 [1] CRAN (R 4.1.2)
#> tensor 1.5 2012-05-05 [1] CRAN (R 4.1.0)
#> testthat 3.1.0 2021-10-04 [1] CRAN (R 4.1.0)
#> tibble 3.2.1 2023-03-20 [1] CRAN (R 4.1.2)
#> tictoc * 1.0.1 2021-04-19 [1] CRAN (R 4.1.0)
#> tidyr 1.3.0 2023-01-24 [1] CRAN (R 4.1.2)
#> tidyselect 1.2.0 2022-10-10 [1] CRAN (R 4.1.2)
#> usethis 2.1.3 2021-10-27 [1] CRAN (R 4.1.0)
#> utf8 1.2.2 2021-07-24 [1] CRAN (R 4.1.0)
#> uwot 0.1.14 2022-08-22 [1] CRAN (R 4.1.2)
#> vctrs 0.6.2 2023-04-19 [1] CRAN (R 4.1.2)
#> vipor 0.4.5 2017-03-22 [1] CRAN (R 4.1.0)
#> viridisLite 0.4.0 2021-04-13 [1] CRAN (R 4.1.0)
#> withr 2.5.0 2022-03-03 [1] CRAN (R 4.1.2)
#> xfun 0.36 2022-12-21 [1] CRAN (R 4.1.2)
#> xtable 1.8-4 2019-04-21 [1] CRAN (R 4.1.0)
#> yaml 2.2.1 2020-02-01 [1] CRAN (R 4.1.0)
#> zoo 1.8-9 2021-03-09 [1] CRAN (R 4.1.0)
#>
#> [1] /Library/Frameworks/R.framework/Versions/4.1/Resources/library
#>
#> ──────────────────────────────────────────────────────────────────────────────