What’s the drug development process?
Has AI changed the course of Drug Development? To answer this question, we need first to understand the drug development process.
The whole process includes the following:
- target identification
- target pharmacology and biomarker development
- lead identification, lead optimization
- Clinical research & development
- regulatory review of IND (investigational new drug) and later phase clinical trials
- post-marketing knowledge
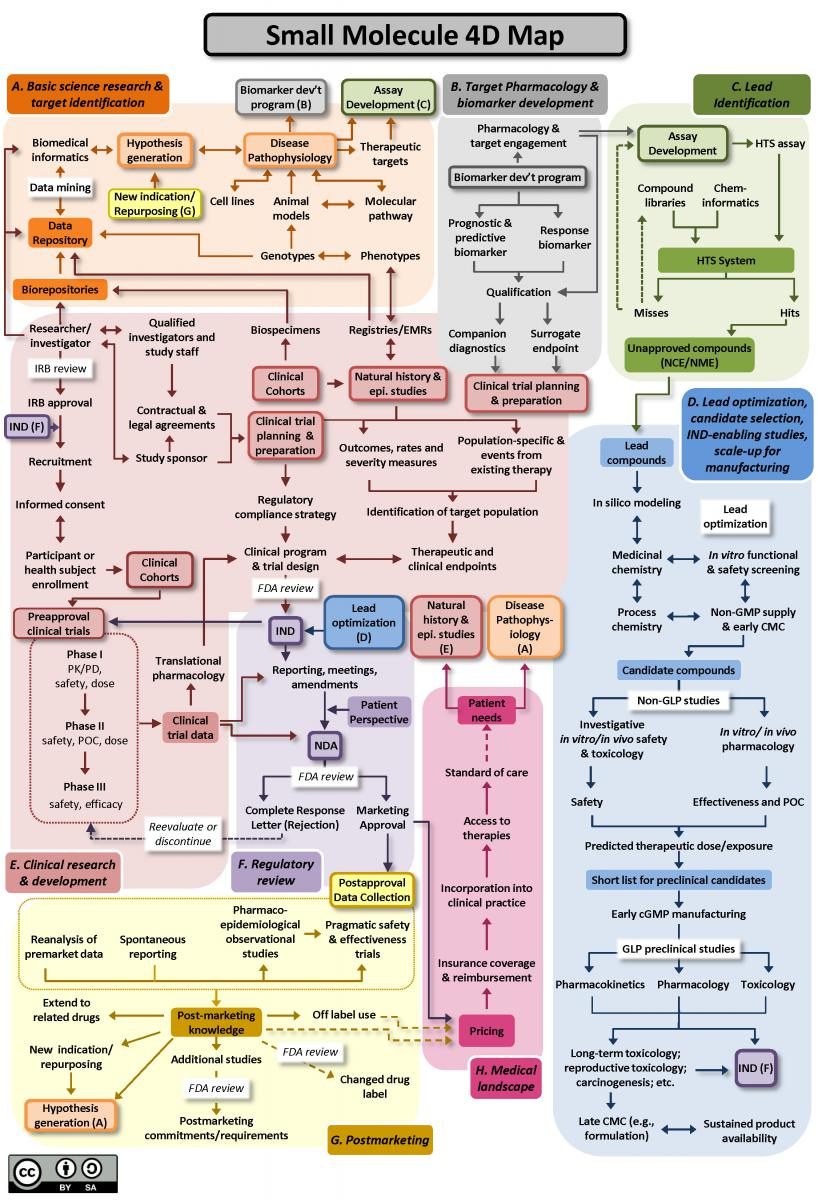
Biologics/antibodies drug development follows a similar path (you can find the map in the same link). As you can see, new target identification is only a tiny component of the complicated drug development process.
What is AI?
AI stands for artificial intelligence. It refers to the development of computer systems or machines that can perform tasks that typically require human intelligence. AI systems are designed to simulate human intelligence and are capable of learning, reasoning, problem-solving, and making decisions.
The definition is broad. I prefer the term machine learning (ML) in the context of bioinformatics which includes: supervised and unsupervised machine learning approaches. This review has a good summary:
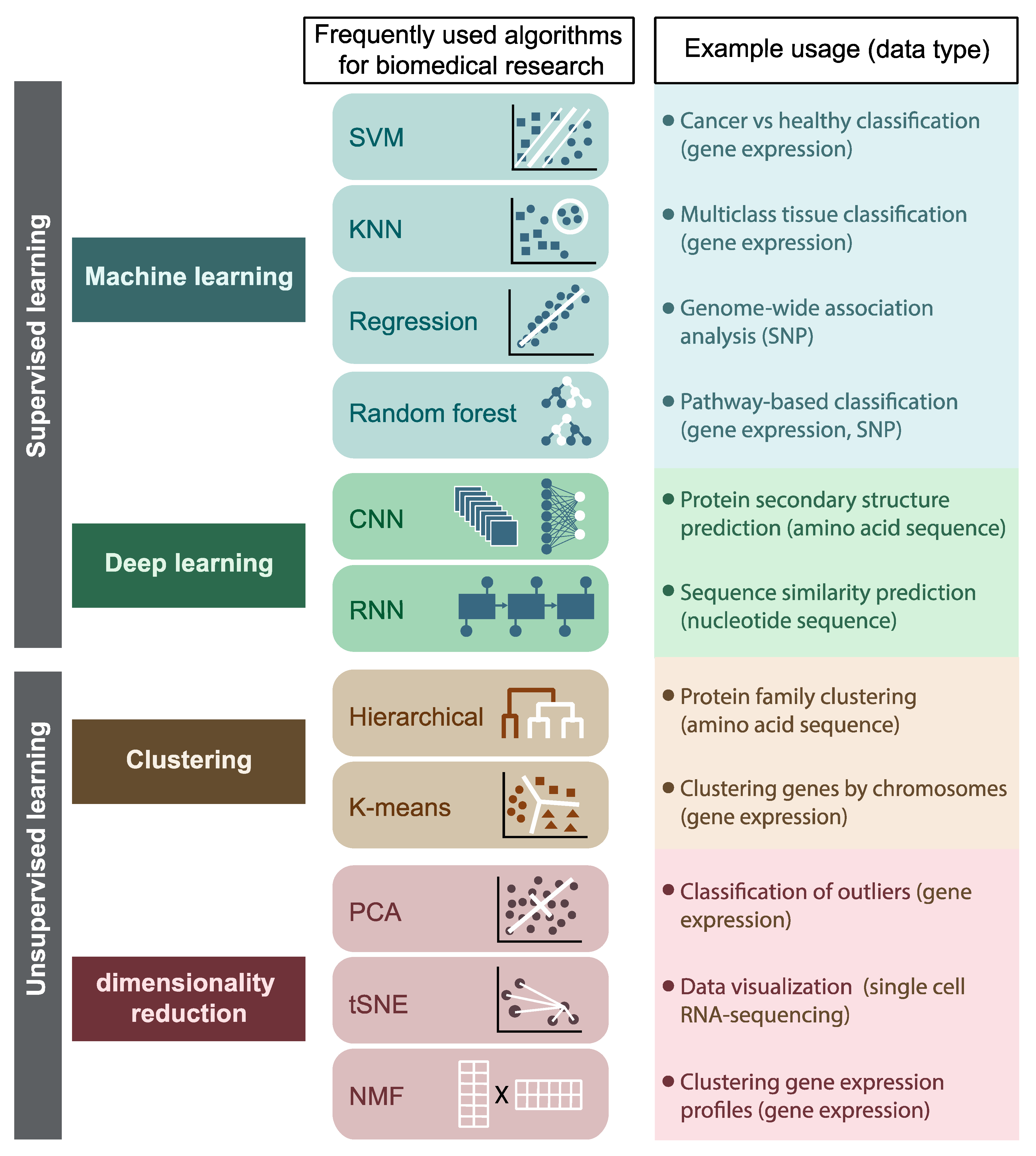
Most AI/ML is applied to target discovery, or the first step of the drug development process. We can mine large of genomics data or single cell data to find new drug targets.
UPDATE on 07/19/2023, I saw this nice figure from https://twitter.com/bwhite5290/status/1680930702039347200 showing what AI can help with the drug development process:
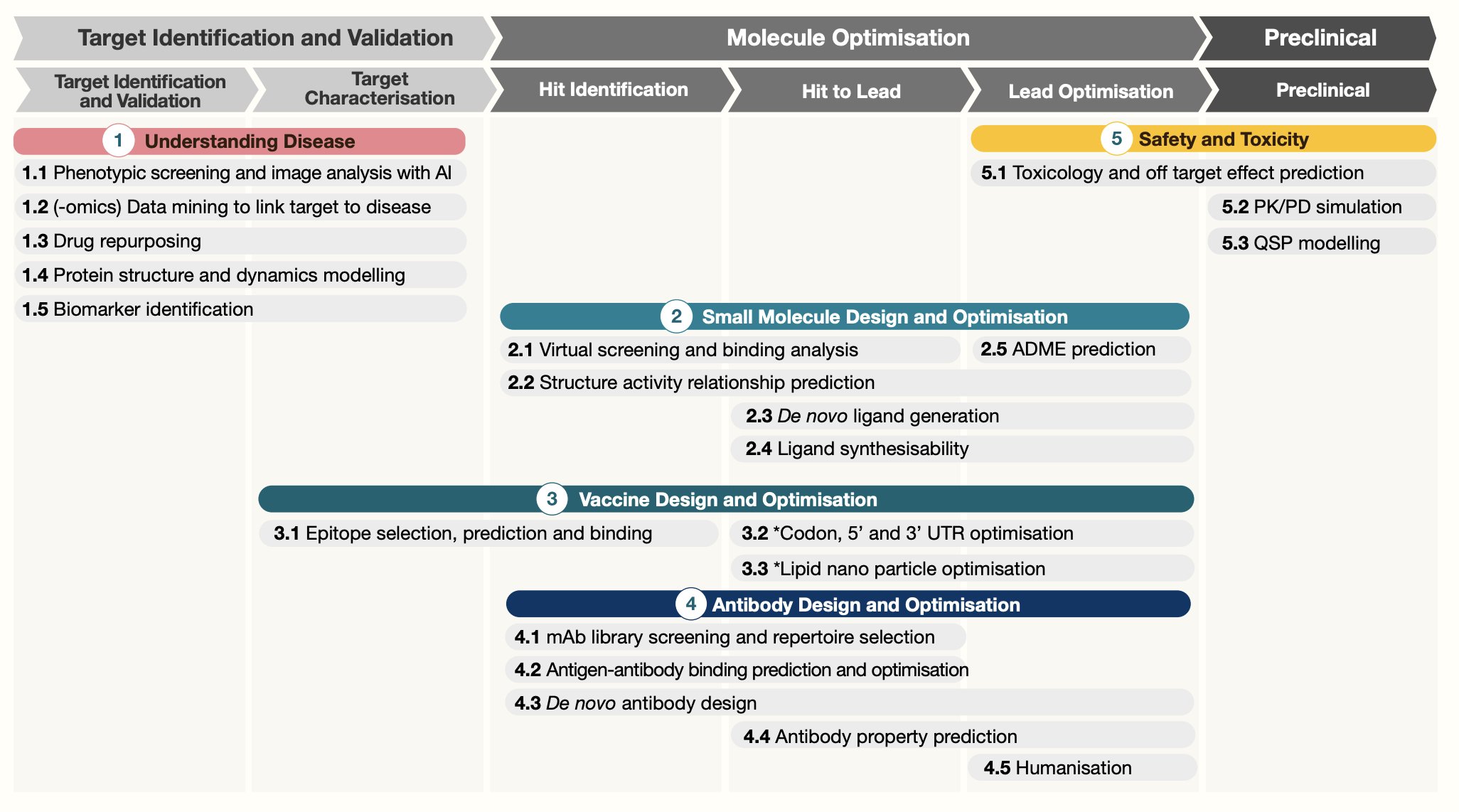
In addition to target identification, we can also apply AI/ML on lead identification and lead optimization. For small molecules, One can screen virtual libraries of large numbers of existing compounds that can inhibit or activate a new target. It can narrow down the list and then validate in experimental assays.
Recursion uses an array of robots to treat millions of cell samples with drugs and genetic perturbations, stain them, and image them. It then applies machine learning algorithms to search for informative relationships between the perturbations and the morphological features of the cells.
Most of the AI companies are vague on their ML approaches in their websites.
My favorite supervised machine learning technique is linear regression (logistic regression for classification) and random forest. They usually perform well when the data size is moderate. In addition, they are simple, easy to interpret compared with deep learning models in which multiple layers are wrapped in a black box. Image analysis and protein structure prediction may be the fields in that deep learning is better than conventional machine learning approaches.
By analyzing single-cell RNAseq, single-cell TCRseq and spatial transcriptome data, our company Immunitas uses conventional machine-learning algorithms (e.g., linear regression and random forest) to identify new targets for immunotherapy. Our approaches do give sensible targets.
In my experience, most of the time, you do not need deep learning; you do not even need machine learning. A simple correlation analysis answers the question in hand.

We are not there yet
With the recent development of the large language model (LLM), the hype of how AI can transform drug development has reached its peak.
What’s the reality?
AlphaFold2 (AF2) from DeepMind has very high accuracy in predicting protein structures. All the structures can be accessed at https://alphafold.ebi.ac.uk/. Can you do antibody discovery with AlphaFold2? In this post:
Experimental conclusion: We performed a very easy experiment to check if there’s “a free lunch” for antibody discovery employing AlphaFold2. Unfortunately, according to our results, this is not the case”
At Immunitas, Matt Bernstein in our group used AlphaFold-multimer for a ligand deorphaning project, and the identified candidates are not validated by the experiment so far.
Derek Lowe wrote,
It is very, very rare for knowledge of a protein’s structure to be any sort of rate-limiting step in a drug discovery project
His comment makes sense as you can see how complicated the whole drug process is from the map in the beginning. Clinical trials usually are the bottlenecks.
AI-designed compounds are still in their early stage. For example, an AI-discovered drug VRG50635 from Verge genomics entered clinical trials. It is announced that the Phase 1 clinical trial data is positive this week. Phase I trial is mainly for safety. Whether it will reach clinical end point in later phases is still yet to see.
Another AI-leading company, BenevolentAI, recently underwent layoffs and restructuring because of their atopic dermatitis program failure in the Phase 2a clinical trial.
Derek Lowe wrote,
There are no existing AI/ML systems that mitigate clinical failure risks due to target choice or toxicology.
Generative AI is being used to design new proteins/antibodies. E.g., @abscibio claims they can de novo design antibodies from scratch, but people disagree. That being said, the field is advancing quickly; see my notes.
See a full list of AI-enabled drugs in clinical trials.
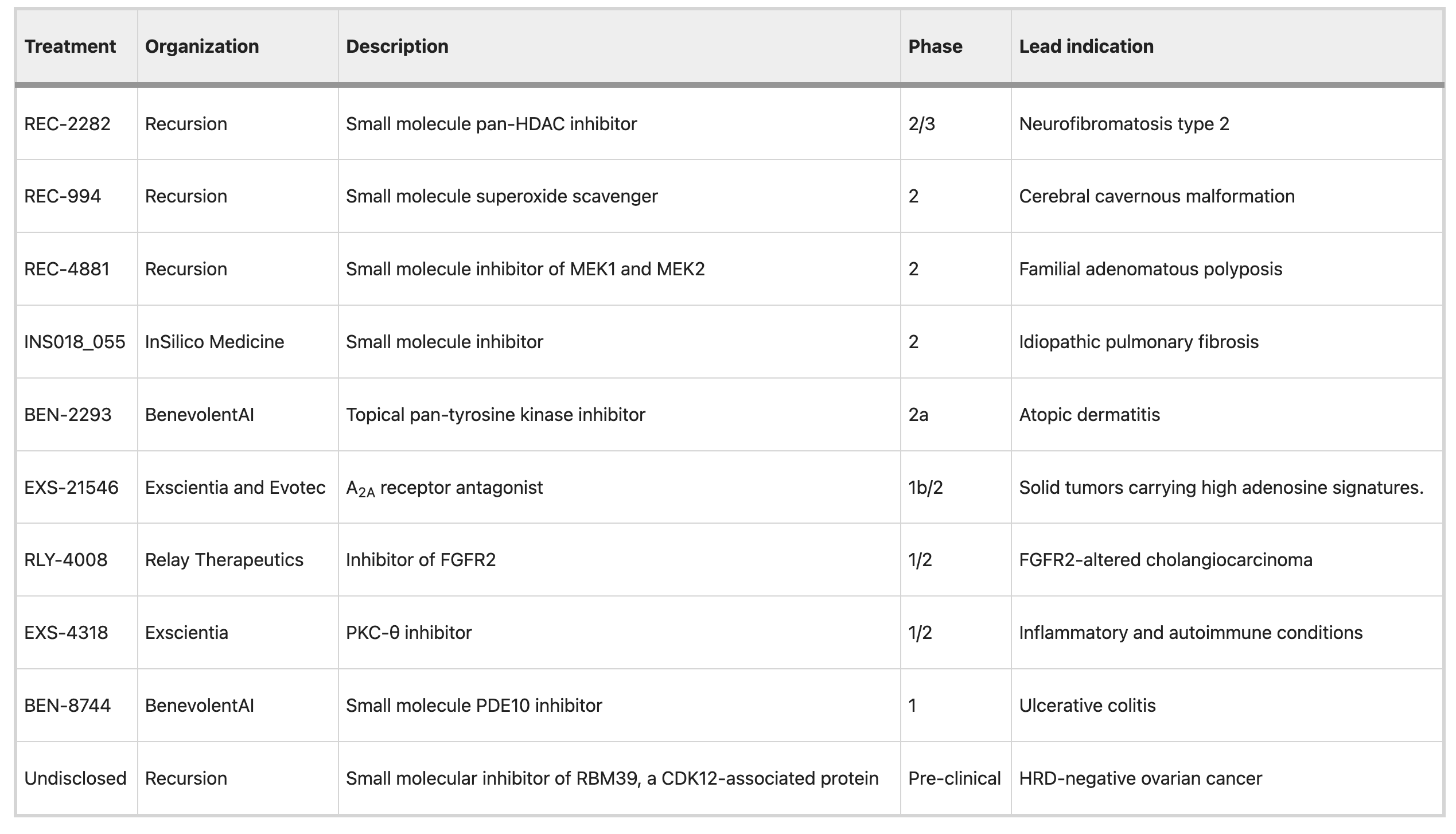
It all gets down to data.
Small molecules are attractive for AI approaches in part owing to the availability of appropriate data to learn from, enabling good predictions about new molecules. Small molecules are well described by their chemical structure, which can be rendered quickly in a format that computers can use.
In order for AI to work, it has to have enough data to train on. But the problem is that we do not have enough high-quality data. Last month, I was in the HubX-Exchange meeting: AI for drug discovery. In the roundtable on data quality which I facilitated, the Chemical informaticians complained about lacking quality data. Genomics data have the same problem. Data silos is another big problem in pharma. Even failed clinical trial data can be valuable but often they are not accessible.
As Jacob mentions in this blog post, the main barrier now is data.
There are two strategies for solving this problem in drug development:
- Better tools and systems for data collection and management (so one can FAIR the data)
- Building around novel high throughput biology (so one can generate a lot of data)
I want to add one more:
- Avoid data silos (so data can be shared and reused)
With enough data, AI could play an important role for drug development. However, we need to be reminded that it is just a tool, like any experimental assay. It is a small piece of the whole puzzle. We still need to perform experiments to validate the AI-discovered/designed drugs and ultimately test them in human patients. With enough high quality data, AI also can help design better clinical trials and recruit the right patients.
Understanding the biology is as important. Bioinformaticians should work closely with biology experts. The domain knowledge is critical in selecting a good target from the top list generated by AI. Even one day, the knowledge graph and LLM are so good, human expertise is still un-replaceable.
AI has not revolutionized drug development yet, but it may first disrupt the early detection of cancer
Two companies, GRAIL and EXAI, announced their liquid biopsy’s high accuracy and specificity for early cancer detection.
This speaks for the success of using AI algorithms for the diagnosis.
References
Tapping into the drug discovery potential of AI https://www.nature.com/articles/d43747-021-00045-7
Artificial intelligence in drug discovery and development https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7577280/
https://decodingbio.substack.com/p/biobyte-032-ai-and-the-overlooked
AI-designed drugs https://www.nature.com/articles/s41591-023-02361-0
https://www.science.org/content/blog-post/has-ai-discovered-drug-now-guess
https://www.digitalisventures.com/blog/engineering-biology-how-to-build-data-centric-biotech
Chris Dawn’s post https://dwan.org/index.php/2023/06/12/a-tale-of-three-conferences/