In this blog post, I am going to show you how to make a correlation heatmap with p-values and significant values labeled in the heatmap body. Let’s use the PBMC single cell data as an example.
You may want to read my previous blog post How to do gene correlation for single-cell RNAseq data.
Load libraries
library(dplyr)
library(Seurat)
library(patchwork)
library(ggplot2)
library(ComplexHeatmap)
library(SeuratData)
library(hdWGCNA)
library(WGCNA)
set.seed(1234)prepare the data
data("pbmc3k")
pbmc3k#> An object of class Seurat
#> 13714 features across 2700 samples within 1 assay
#> Active assay: RNA (13714 features, 0 variable features)## routine processing
pbmc3k<- pbmc3k %>%
NormalizeData(normalization.method = "LogNormalize", scale.factor = 10000) %>%
FindVariableFeatures(selection.method = "vst", nfeatures = 2000) %>%
ScaleData() %>%
RunPCA(verbose = FALSE) %>%
FindNeighbors(dims = 1:10, verbose = FALSE) %>%
FindClusters(resolution = 0.5, verbose = FALSE) %>%
RunUMAP(dims = 1:10, verbose = FALSE)
Idents(pbmc3k)<- pbmc3k$seurat_annotations
pbmc3k<- pbmc3k[, !is.na(pbmc3k$seurat_annotations)]construct metacell
pbmc3k <- SetupForWGCNA(
pbmc3k,
gene_select = "fraction", # the gene selection approach
fraction = 0.05, # fraction of cells that a gene needs to be expressed in order to be included
wgcna_name = "tutorial" # the name of the hdWGCNA experiment
)
# construct metacells in each group.
# choosing K is an art.
pbmc3k <- MetacellsByGroups(
seurat_obj = pbmc3k,
group.by = c("seurat_annotations", "orig.ident"), # specify the columns in seurat_obj@meta.data to group by
k = 10, # nearest-neighbors parameter
max_shared = 5, # maximum number of shared cells between two metacells
ident.group = 'seurat_annotations' # set the Idents of the metacell seurat object
)
# extract the metacell seurat object
pbmc_metacell <- GetMetacellObject(pbmc3k)
pbmc_metacell@meta.data %>% head()#> orig.ident nCount_RNA nFeature_RNA
#> B#pbmc3k_1 pbmc3k 1794.8 3237
#> B#pbmc3k_2 pbmc3k 4170.4 4962
#> B#pbmc3k_3 pbmc3k 1765.7 3233
#> B#pbmc3k_4 pbmc3k 1731.3 3060
#> B#pbmc3k_5 pbmc3k 1982.2 3468
#> B#pbmc3k_6 pbmc3k 3084.5 4351
#> cells_merged
#> B#pbmc3k_1 CGCCATTGGAGCAG,TGCCGACTCTCCCA,GGTACATGAAAGCA,GATTGGACGGTGTT,ATCCCGTGGCTGAT,CCAGCTACCAGCTA,TTGGAGACTATGGC,GAAGTCACCCTGTC,CGAGGGCTACGACT,TTATCCGACTAGTG
#> B#pbmc3k_2 AAACATTGAGCTAC,TATGTCACGGAACG,AAACTTGAAAAACG,GGAGCCACCTTCTA,CATTTGTGGGATCT,GCTAGAACAGAGGC,CAGGTTGAGGATCT,ACTGAGACGTTGGT,ACGAAGCTCTGAGT,CGCGAGACAGGTCT
#> B#pbmc3k_3 CTACTATGTAAAGG,CCAGGTCTACACCA,TTGGTACTGAATCC,ATCGCGCTTTTCGT,GCACTAGAAGATGA,ATGACGTGACGACT,GGGCCAACGCGTTA,CGCCATTGGAGCAG,GAAAGCCTACGTTG,CATCATACGGAGCA
#> B#pbmc3k_4 GAGAAATGTTCTCA,ACCCAAGATTCACT,GGGCCAACGCGTTA,GAGTGGGAGTCTTT,TATTGCTGCCGTTC,GGCAATACGCTAAC,CGAGGGCTACGACT,CAGTGATGTACGCA,TCGATTTGTCGTGA,TGATCACTTCTACT
#> B#pbmc3k_5 CAGTTACTAAGGTA,AAGTGGCTTGGAGG,TGATTCACTGTCAG,TGCGAAACAGTCAC,TATACCACCTGATG,TTATTCCTGGTACT,TTGACACTGATAAG,CGAGGGCTACGACT,TACTCAACTGCTAG,TAGCTACTGAATAG
#> B#pbmc3k_6 TTCCAAACCTCCCA,CGCGATCTTTCTTG,AAATCAACAATGCC,ATCCCGTGGCTGAT,ACGAAGCTCTGAGT,TTTAGAGATCCTCG,AAACATTGAGCTAC,ACGACCCTTGACAC,GTAGCAACCATTTC,GAATTAACTGAAGA
#> seurat_annotations
#> B#pbmc3k_1 B
#> B#pbmc3k_2 B
#> B#pbmc3k_3 B
#> B#pbmc3k_4 B
#> B#pbmc3k_5 B
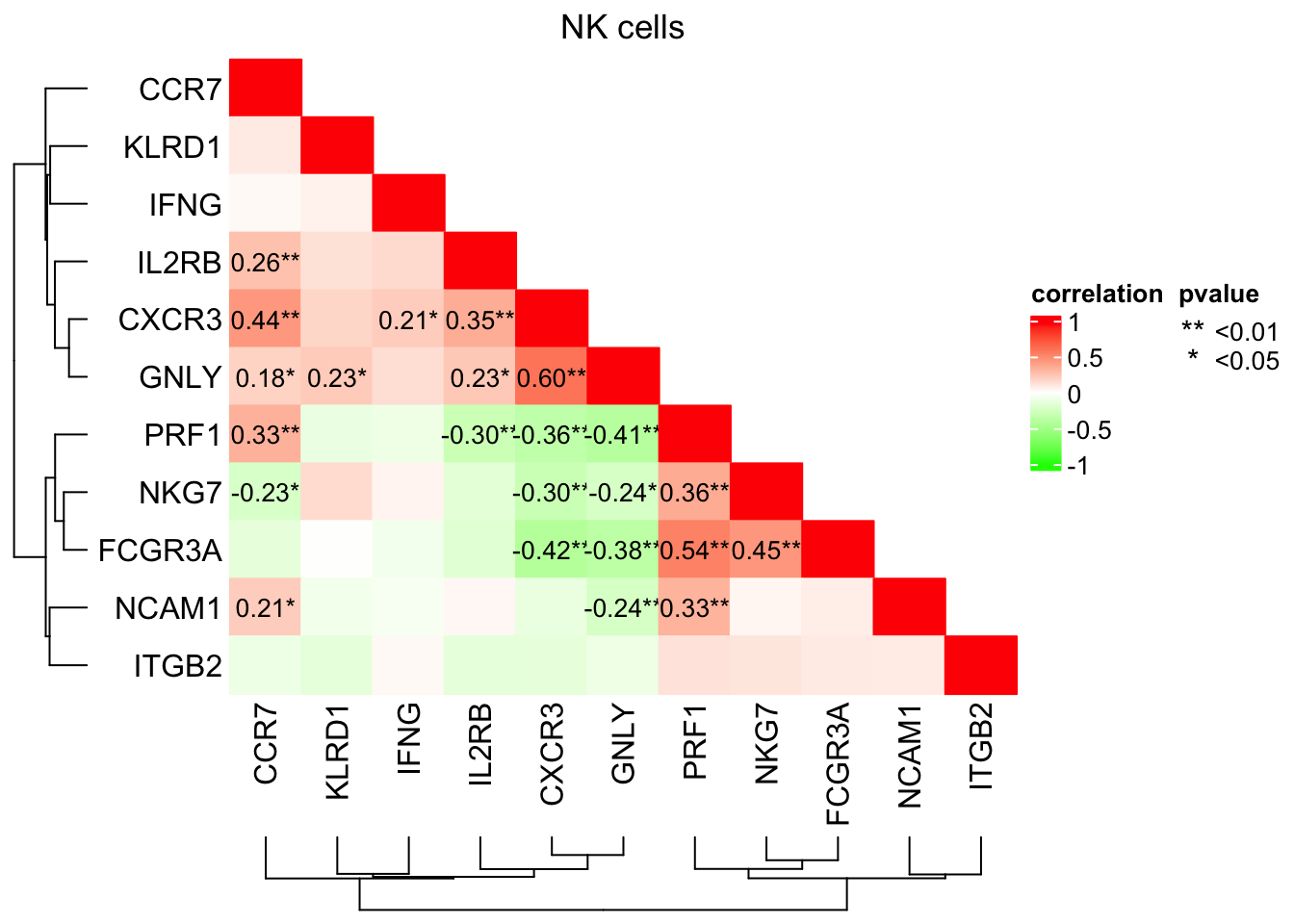
#> B#pbmc3k_6 BHmisc::rcorr function gives the pair-wise correlation coefficient and p-value
genes<- c("GNLY", "NKG7", "KLRD1", "ITGB2", "PRF1", "IFNG", "NCAM1", "FCGR3A", "CCR7", "CXCR3", "IL2RB")
cells<- WhichCells(pbmc_metacell, expression = seurat_annotations == "NK")
mat<- pbmc_metacell@assays$RNA@data[genes, cells] %>%
as.matrix() %>%
t()
# Hmisc package gives the pair-wise correlation coefficient and p-value
cor_res<- Hmisc::rcorr(mat)
cor_mat<- cor_res$r
# sometimes, you may have NA in the matrix, and clustering does not play well with it
# a simple hack is to turn the NA to 0
cor_mat[is.na(cor_mat)]<- 0
cor_p<- cor_res$P
cor_p#> GNLY NKG7 KLRD1 ITGB2 PRF1 IFNG
#> GNLY NA 1.034720e-02 0.01446758 0.2740351 5.598974e-06 0.13474999
#> NKG7 1.034720e-02 NA 0.09914756 0.2480082 7.635844e-05 0.59502786
#> KLRD1 1.446758e-02 9.914756e-02 NA 0.1174652 1.951340e-01 0.54200187
#> ITGB2 2.740351e-01 2.480082e-01 0.11746519 NA 1.956304e-01 0.77673864
#> PRF1 5.598974e-06 7.635844e-05 0.19513395 0.1956304 NA 0.27047205
#> IFNG 1.347500e-01 5.950279e-01 0.54200187 0.7767386 2.704720e-01 NA
#> NCAM1 8.662654e-03 6.572153e-01 0.39486873 0.3767390 3.435796e-04 0.60012618
#> FCGR3A 2.259989e-05 4.024618e-07 0.92716018 0.3407721 3.353928e-10 0.45453683
#> CCR7 4.702197e-02 1.452948e-02 0.34889863 0.2674199 2.461824e-04 0.78811106
#> CXCR3 9.232615e-13 8.725519e-04 0.06138810 0.1045795 6.629722e-05 0.02079542
#> IL2RB 1.199037e-02 7.699923e-02 0.16780117 0.1016665 9.815965e-04 0.09288220
#> NCAM1 FCGR3A CCR7 CXCR3 IL2RB
#> GNLY 0.0086626537 2.259989e-05 4.702197e-02 9.232615e-13 0.0119903711
#> NKG7 0.6572153099 4.024618e-07 1.452948e-02 8.725519e-04 0.0769992304
#> KLRD1 0.3948687306 9.271602e-01 3.488986e-01 6.138810e-02 0.1678011737
#> ITGB2 0.3767389762 3.407721e-01 2.674199e-01 1.045795e-01 0.1016664987
#> PRF1 0.0003435796 3.353928e-10 2.461824e-04 6.629722e-05 0.0009815965
#> IFNG 0.6001261787 4.545368e-01 7.881111e-01 2.079542e-02 0.0928821980
#> NCAM1 NA 4.384662e-01 2.119770e-02 1.787253e-01 0.7278303881
#> FCGR3A 0.4384662288 NA 1.065590e-01 1.939171e-06 0.0523142134
#> CCR7 0.0211977006 1.065590e-01 NA 5.449032e-07 0.0040966177
#> CXCR3 0.1787252622 1.939171e-06 5.449032e-07 NA 0.0001233260
#> IL2RB 0.7278303881 5.231421e-02 4.096618e-03 1.233260e-04 NA## add significant **
cor_p[is.nan(cor_p)]<- 1
## the diagonal are NA, make them 1
cor_p[is.na(cor_p)]<- 1
col_fun<- circlize::colorRamp2(c(-1, 0, 1), c("green", "white", "red"))Only lable the correlation coefficients with p-values that are <=0.05; add * for p value <=0.05 and ** for p value <=0.01
cell_fun = function(j, i, x, y, w, h, fill){
if(as.numeric(x) <= 1 - as.numeric(y) + 1e-6) {
grid.rect(x, y, w, h, gp = gpar(fill = fill, col = fill))
}
if (cor_p[i, j] < 0.01 & as.numeric(x) <= 1 - as.numeric(y) + 1e-6){
grid.text(paste0(sprintf("%.2f", cor_mat[i, j]),"**"), x, y, gp = gpar(fontsize = 10))
} else if (cor_p[i, j] <= 0.05 & as.numeric(x) <= 1 - as.numeric(y) + 1e-6){
grid.text(paste0(sprintf("%.2f", cor_mat[i, j]),"*"), x, y, gp = gpar(fontsize = 10))
}
}
hp<- ComplexHeatmap::Heatmap(cor_mat,
rect_gp = gpar(type = "none"),
column_dend_side = "bottom",
column_title = "NK cells",
name = "correlation", col = col_fun,
cell_fun = cell_fun,
cluster_rows = T, cluster_columns = T,
row_names_side = "left")
lgd_list = list(
Legend( labels = c("<0.01", "<0.05"), title = "pvalue",
graphics = list(
function(x, y, w, h) grid.text("**", x = x, y = y,
gp = gpar(fill = "black")),
function(x, y, w, h) grid.text("*", x = x, y = y,
gp = gpar(fill = "black")))
))
draw(hp, annotation_legend_list = lgd_list, ht_gap = unit(1, "cm") )