To not miss a post like this, sign up for my newsletter to learn computational biology and bioinformatics.
In my last blog post, I talked about some common bioinformatics mistakes.
Today, we are going to talk about THE MOST common bioinformatics mistake people make. And I think it deserves a separate post about it. Even some experienced programmers get it wrong and the mistake prevails in many bioinformatics software:
The one-off mistake!!
What is that?
Genomics files such as bed, vcf, gtf etc come with a coordinate system. To describe a region a position in a genome, you specify something like chr3, 4-7. This means the region is on chromosome 3 and from base 4 to 7. But there are more nuances here..
There are the 0-based and 1-based coordinates systems in genomics:
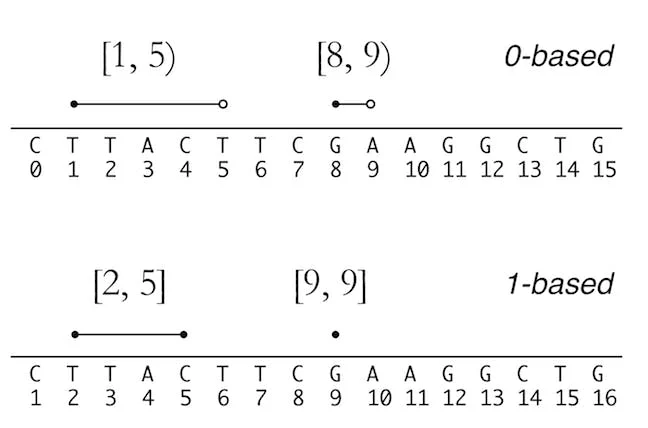
The 0-based starts with 0 and the 1-based begins with 1. Why it matters?
Different file formats (THE bioiFORMATics problem!!) use different systems:
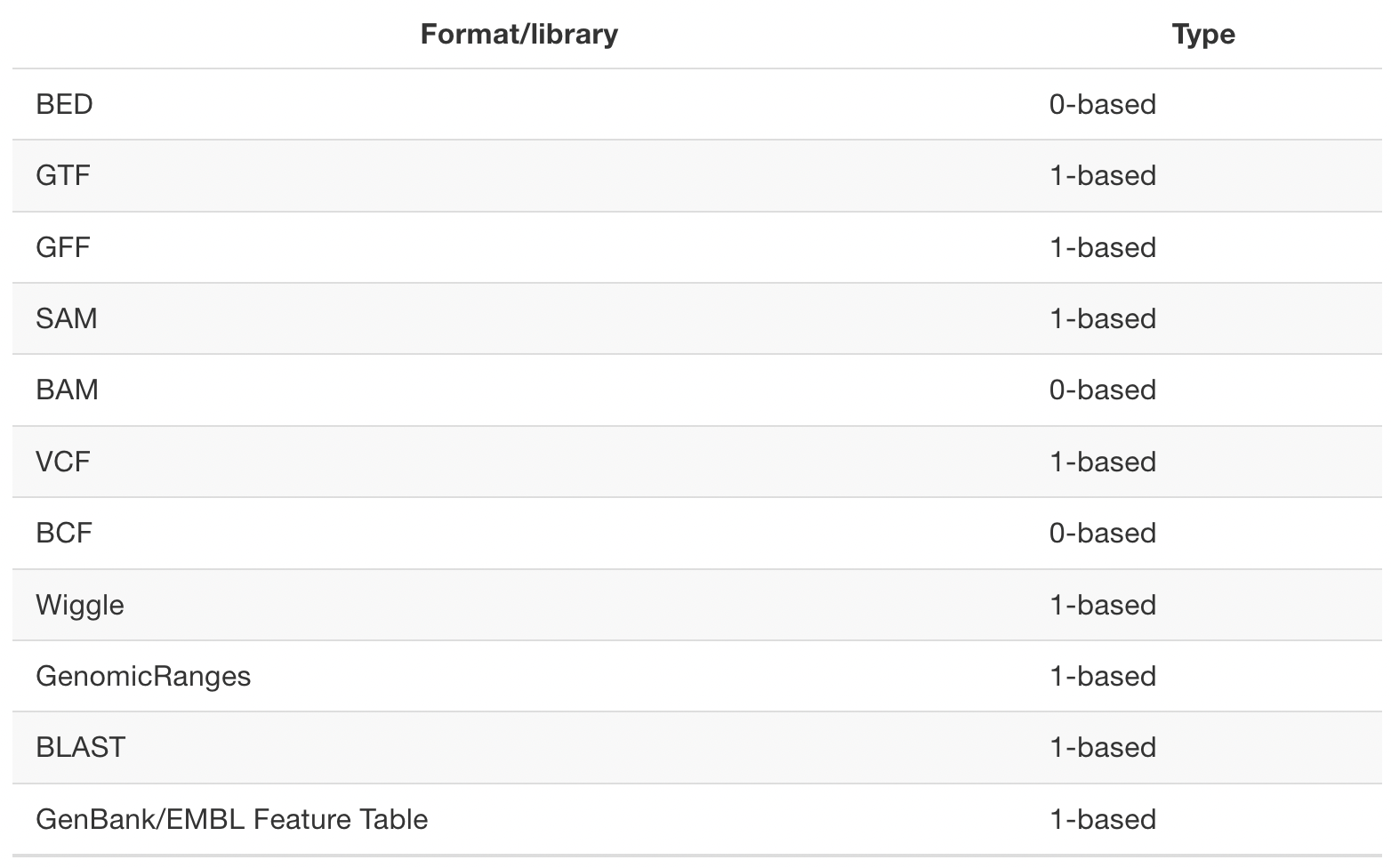
So if you are not careful, you will make one-off mistakes.
I highly recommend you to read this old post on biostars: https://www.biostars.org/p/84686/
To make things worse :) you know R is 1-based and Python is 0-based!!
If you use different languages to analyze the data, you must also be careful.
some files such as bed file is 0 based. Two genomic regions:
chr1 0 1000
chr1 1000 2000when you import that bed file into R using rtracklayer::import(), it will become
chr1 1 1000
chr1 1001 2000The function converts it to 1 based internally (R is 1 based unlike Python).
When you read the bed file with read.table and use GenomicRanges::makeGRangesFromDataFrame() to convert it to a GRanges object, do not forget to add 1 to the start before doing it.
Similarly, when you write a GRanges object to disk using rtracklayer::export, you do not need to worry, R will convert it back to 0 based in the file. However, if you make a dataframe out of the GRanges object, and write that dataframe to file, remember to do start -1 before doing it.
I hope you learned something new!
Further reading: UCSC coordinate counting system https://genome-blog.gi.ucsc.edu/blog/2016/12/12/the-ucsc-genome-browser-coordinate-counting-systems/
Happy Learning!
Tommy