 My caption 😄
My caption 😄
Synthetic essentiality of chromatin remodelling factor CHD1 in PTEN-deficient cancer
Abstract
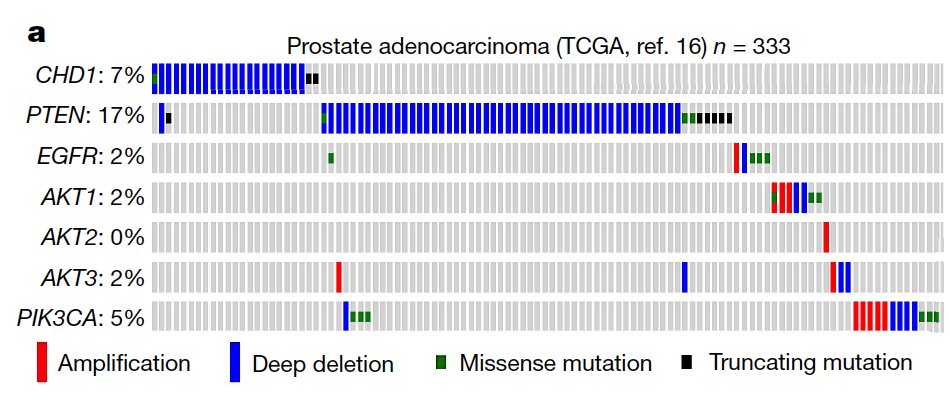
Synthetic lethality and collateral lethality are two well-validated conceptual strategies for identifying therapeutic targets in cancers with tumour-suppressor gene deletions1–3. Here, we explore an approach to identify potential synthetic-lethal interactions by screening mutually exclusive deletion patterns in cancer genomes. We sought to identify ‘synthetic-essential’ genes: those that are occasionally deleted in some cancers but are almost always retained in the context of a specific tumour-suppressor deficiency. We also posited that such synthetic-essential genes would be therapeutic targets in cancers that harbour specific tumour-suppressor deficiencies. In addition to known synthetic-lethal interactions, this approach uncovered the chromatin helicase DNA-binding factor CHD1 as a putative synthetic-essential gene in PTEN-deficient cancers. In PTEN-deficient prostate and breast cancers, CHD1 depletion profoundly and specifically suppressed cell proliferation, cell survival and tumorigenic potential. Mechanistically, functional PTEN stimulates the GSK3β-mediated phosphorylation of CHD1 degron domains, which promotes CHD1 degradation via the β-TrCP-mediated ubiquitination–proteasome pathway. Conversely, PTEN deficiency results in stabilization of CHD1, which in turn engages the trimethyl lysine-4 histone H3 modification to activate transcription of the pro-tumorigenic TNF–NF-κB gene network. This study identifies a novel PTEN pathway in cancer and provides a framework for the discovery of ‘trackable’ targets in cancers that harbour specific tumour-suppressor deficiencies.
More detail can easily be written here using Markdown and $\rm \LaTeX$ math code.